 EHR have been increasingly used for research purposes due to the tremendous depth of patient data available, and the extensive information on health outcomes and risk factors contained in them. This tremendous trove of patient data necessitates the advancement of automated high-throughput phenotyping algorithms to expedite the identification of relevant patient cohorts from EHRs with clearly defined phenotypes. The resulting EHR-derived phenotypes are then used for general purpose knowledge discovery, including identification of risk factors for chronic conditions, evaluation of efficacy for new treatment options, prediction of adverse events following drug usage, drug repurposing, discovery of drug-drug interactions, and many others. While EHRs have been used for phenotyping and disease-related data mining for several years, there remain some major challenges to this type of research. A key challenge remains that the reproducibility of findings across studies is limited, which raised a fundamental concern on the value of these researching findings.
EHR have been increasingly used for research purposes due to the tremendous depth of patient data available, and the extensive information on health outcomes and risk factors contained in them. This tremendous trove of patient data necessitates the advancement of automated high-throughput phenotyping algorithms to expedite the identification of relevant patient cohorts from EHRs with clearly defined phenotypes. The resulting EHR-derived phenotypes are then used for general purpose knowledge discovery, including identification of risk factors for chronic conditions, evaluation of efficacy for new treatment options, prediction of adverse events following drug usage, drug repurposing, discovery of drug-drug interactions, and many others. While EHRs have been used for phenotyping and disease-related data mining for several years, there remain some major challenges to this type of research. A key challenge remains that the reproducibility of findings across studies is limited, which raised a fundamental concern on the value of these researching findings.
We are currently developing a framework of prior-knowledge-guided integrated likelihood model with readily available software to account for the EHR data error from imperfect phenotyping algorithms, which will ultimately enhance the reproducibility of EHR based discovery; see the figure below.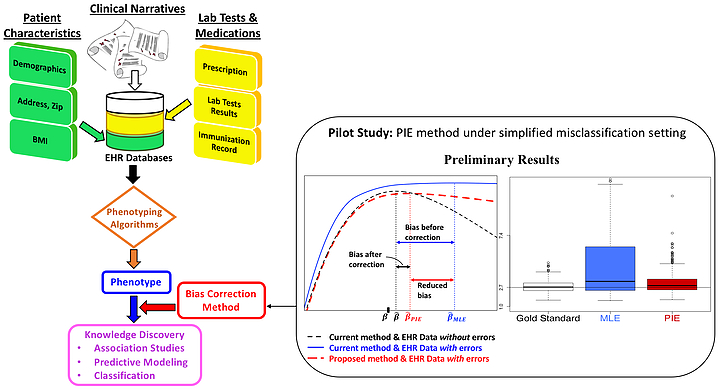
In the past, we have been developing novel methods to account for errors in EHR data. In our paper presented at the AMIA 2016 Annual Symposium, we used extensive simulation studies guided by eMERGE (Electronic Medical Records and Genomics) data to quantify the loss of power due to different levels of phenotyping errors in association studies (Duan et al., 2016, AMIA Annual Symposium Proceedings). This paper won the first prize of 2016 \Best of Student Papers in Knowledge Discovery and Data Mining (KDDM)”Awards in AMIA 2016.
More recently, we have developed an innovative statistical method using the cutting-edge theory of integrated likelihood to correct for the estimation bias caused by phenotyping errors (Huang et al., 2017, JAMIA); see the figure below.
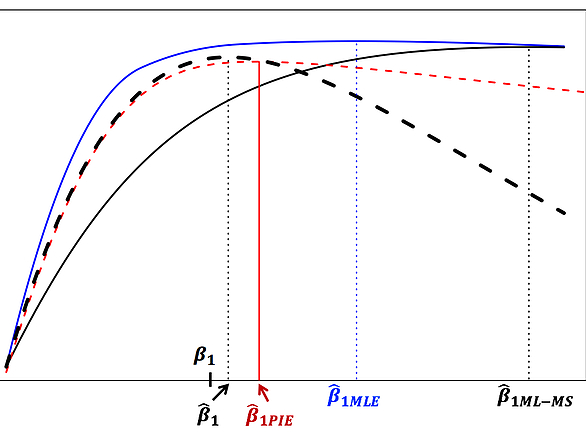
We are currently expanding this framework to tackle more realistic settings of differential misclassifications in EHR-derived phenotypes.
